Sabtu, 14 April 2012 merupakan salah satu hari penting di JSC. WHY??? karena dihari itu diadakannya Upgrading JSC yang merupakan program dari Biro PSDM dengan TIK mempererat kekeluargaan antara fungsionaris JSC.
Halaman Auditorium.....
ya, di sanalah tempat diadakannya upgrading JSC pukul 07.00-11.00 WIB. Kegiatan yang dihadiri oleh kurang lebih 15 dari 33 fungsionaris JSC ini diawali dengan sambutan dari ketua JSC saudara Bambang yang memberi semangat kepada fungsionaris yang datang saat itu.
Kurang lengkap bila upgrading tanpa games. Tentunya salah satu capaian Upgrading yaitu have fun/merefreskan diri dari segelintir tugas kuliah. Nah gamesnya apa aja? ada Estafet Balon, dan Nyanyi ala Pas Mantap. Pokoknya seru walaupun kedatangan fungsionaris tidak lengkap.
Acara terakhir yaitu para fungsionaris menuliskan pesan dan kesannya selama di JSC dan Upgrading hari itu, kemudian beberapanya dibacakan.
Semoga JSC dapat mempererat kekeluargaan antara semua fungsionaris sehingga banyak fungsionaris yang datang dalam kegiatan JSC.
JSC!!! Be The Winner (4x)
Minggu, 15 April 2012
Jumat, 06 April 2012
Big pest, small genome

Scanning electron microscope image of a two-spotted spider mite, which is less than one millimeter long. A University of Utah biologist and an international research team decoded the genetic blueprint of the two-spotted spider mite, raising hope for new ways to attack the major pest, which resists pesticides and destroys crops and ornamental plants worldwide.
The voracious mites, which technically are not insects, can eat more than 1,100 plant species – a rare trait. The mites' newly revealed and sequenced genome contains a variety of genes capable of detoxifying pesticides as well as toxins plants use to defend themselves, the scientists report in the Thursday, Nov. 24 issue of the journal Nature.
"One key thing that makes spider mites unique is they can eat many, many different plant species," says Richard M. Clark, one of five main authors of the study and an assistant professor of biology at the University of Utah. "These mites are often house plant pests – a major cause of people's house plants turning yellow and getting sick. They also are a major problem for agricultural nurseries and greenhouses, and for field crops." Primary targets are tomatoes, peppers, cucumbers, strawberries, corn, soybeans, apples, grapes and citrus.
Clark says the new study's "importance is largely in understanding how animals eat plants, with the long-term goal of developing effective ways to prevent crop damage from mites and insects. If we can identify the biological pathways mites use to feed on plants, we can potentially identify chemical and biological methods to disrupt those pathways and stop the mites from feeding."
The two-spotted spider mite, which is no more than 1 millimeter long, "is a major global pest, and is predicted to be a growing concern in a warming climate because they multiply extremely fast at high temperatures – 90 degrees Fahrenheit or more," he adds. "They do really well in hot and dry climates like Utah." __IMAGE_2
Yet, the two-spotted spider mite "has been found to rapidly develop resistance to multiple types of pesticides, often within a couple of years after a pesticide is introduced," says Clark. "It is resistant to many common pesticides used against insects."
The Nature study deciphering the genome of Tetranychus urticae, the two-spotted spider mite (which has two red spots), was conducted by an international research team of 55 scientists from North America, Europe and South America.
Besides Clark, the other primary authors are biologists Yves Van de Peer of Ghent University and the Flanders Institute for Biotechnology in Belgium; Miodrag Grbic of the University of Western Ontario, Canada; Thomas Van Leeuwen of Ghent University; and Rene Feyereisen of the University of Nice Sophia Antipolis in France.
Genetic Blueprint of the Two-Spotted Spider Mite
Decoding the spider mite's genome required dozens of scientists with expertise in various gene families. Clark mainly studied which genes are "expressed" or activated and thus make messenger RNA, or mRNA, which in turn is used to make proteins.
The study found that the two-spotted spider mite has 18,414 genes. Clark and University of Utah graduate student Edward J. Osborne found that 15,397 genes are "expressed" or activated to make proteins. __IMAGE_3
The spider mite genome contains about 90 megabases – that's 90 million "base pairs" of DNA letters (A,C, G and T) – which is the smallest genome yet sequenced for any arthropod, which are invertebrate or spineless animals with external skeletons or exoskeletons, segmented bodies, and appendages with joints.
"Many of the other genomes are enormous," some close to 3 billion bases, or about the size of the human genome, and some up to 7.1 billion bases, Clark says.
Arthropods include hexapods (insects and insect-like animals), crustaceans (lobsters, crabs, shrimp, barnacles), myriapods (millipedes, centipedes) and chelicerates (spiders, scorpions, mites and ticks). Chelicerates are the largest group of animals after insects. The two-spotted spider mite is the first chelicerate to have its genome fully sequenced.
While there are other species of plant-feeding mites, the researchers chose to sequence the genome of the two-spotted spider mite "because of all the spider mites, this is the most widespread because it feeds on so many different plant species," Clark says.
The study's findings shed light on how the spider mites evolved differently than other arthropods. Compared with other arthropods, the two-spotted spider mite:
- Uses a different molting hormone to shed its exoskeleton during growth.
- Has only eight Hox genes to orchestrate body-plan development, compared with 10 in most other arthropods, and thus has only two main body segments instead of three. There are other cases in which Hox genes were activated differently in different arthropods, "but this is an extreme case," Clark says. "The genes are both gone.
- Makes silk that is strong like spider silk but 185 to 435 times thinner. "Spiders spin silk from their abdomens, spider mites from the head region," Clark says. Spider mites use silk to hide from predators, keep themselves warm, and suspend eggs out of predators' reach. Silk from the mites may prove useful as biodegradable surgical sutures and bandages. It "is very thin and very easy to get because you can grow lots of mites on plants," Clark says.
The spider mite genome also revealed the presence of "families of genes involved in breaking down toxic compounds, either in plants poisonous to the spider mites or in pesticides," says Clark. "You would imagine that if these mites feed on such a broad number of plant hosts, they would have many genes known to be involved in breaking down toxic compounds. And we found that they did."
In some specific families of detoxification genes in the spider mites, "the number of genes was about three times that seen in other arthropods," he adds.
As part of the study, the scientists took a specific strain of spider mites that normally eat kidney beans and transferred them to tomato and thale cress (Arabidopsis) plants. On these new plants, the mites "expressed" or activated different genes and thus made different detoxification compounds so they could eat the new plant species. Some of those detox genes were previously unknown and thus provide new insight into how mites counteract plant defenses.
For example, half of the cytochrome P450 family of detoxification genes changed expression – turned either on or off – when the mites were switched to the new plants. That is a bigger change than seen before in any group of animals, Clark says.
"This suggests that these genes are critical for the ability of mites to be pests on many different plants."
Clark says the spider mite has 39 genes from one drug-resistance gene family (and the proteins they encode), compared with only nine to 14 in insects and vertebrate animals. That shows how an expanded set of genes evolved to help the pests feed on numerous plant species.
In a yet-unexplained mystery, the two-spotted spider mite has some genes similar to those in bacteria and fungi. "They somehow captured them from other organisms in the environment and now are using them for their own growth and persistence," says Clark. "They are mostly enzymes involved in changing other small molecules. The hypothesis is these genes may be involved in modifying [detoxifying] toxic compounds found in plants." Source : University of Utah
http://www.biologynews.net/archives/2011/11/23/big_pest_small_genome.html
New immune defense enzyme discovered
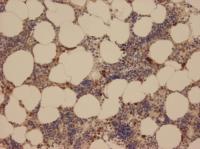
This is a microscope image of normal human bone marrow tissue with stained NSP4 in myeloblasts and myelocytes. Neutrophil granulocytes comprise important defences for the immune system. When pathogenic bacteria penetrate the body, they are the first on the scene to mobilise other immune cells via signal molecules, thereby containing the risk. To this end, they release serine proteases – enzymes that cut up other proteins to activate signal molecules. Scientists at the Max Planck Institute of Neurobiology in Martinsried have now discovered a new serine protease: neutrophil serine protease 4, or NSP4. This enzyme could provide a new target for the treatment of diseases that involve an overactive immune system, such as rheumatoid arthritis.
The functioning of the immune system is based on the complex interplay of the most diverse cells and mediators. For example, neutrophil granulocytes (a group of specialized white blood cells) react to bacteria by releasing substances called serine proteases. These enzymes are able to activate signal molecules, such as the chemokines, by cleaving them at a specific position on the molecule. The active signal molecules then guide other immune cells to the focus of inflammation in order to destroy the pathogens.
A research team led by Dieter Jenne at the Max Planck Institute of Neurobiology in Martinsried has come across a previously unknown protease in humans: neutrophil serine protease 4, or NSP4. "The special thing about this enzyme is that it cuts proteins that have the amino acid arginine at a particular point", says Dieter Jenne, research group leader at the Martinsried-based Institute. "This is where NSP4 differs from the other three known neutrophil serine proteases, which are similar in molecular structure, but have a different recognition motif." The scientists may be able to harness this difference to develop an active substance that specifically inhibits NSP4, thereby reducing the immune reaction. However, serine protease activity comes at a cost. The enzymes not only heal inflammations, but sometimes cause them in the first place. If too many immune cells are activated, they can use their arsenal of aggressive chemical weapons against the body's own tissues. A number of chronic inflammatory diseases are based on precisely this effect. As a result, scientists are searching for substances that can block the neutrophil proteases. To date, however, none of the substances tested have been developed into effective drugs.
"So far, we don't know the identity of the NSP4 substrate, but we assume they must be signal molecules", says Dieter Jenne. Activated chemokines can recruit a vast number of neutrophils, and their sheer quantity alone is enough to cause tissue damage. "Proteases sometimes act as accelerants and can even trigger a chronic inflammation quite independently of bacterial intruders. If we dampened down the defences, we could counteract this effect", explains the scientist.
In terms of evolutionary history, NSP4 is the oldest of the four known neutrophil serine proteases. Using gene sequences, scientists have shown that the enzyme has hardly changed through hundreds of millions of years of evolution from bony fish to humans. "That would indicate that NSP4 regulates a fundamental process", says Dieter Jenne.
The fact that the enzyme remained undiscovered until now is because it occurs at a much lower concentration than the other three proteases. The Max Planck scientists came across it while searching the human genome for genes that encode serine proteases. In the process, they noticed a previously unknown gene sequence. Natascha C. Perera, a member of the Martinsried research group and lead author of the study, managed to produce and examine the enzyme in its active, folded state.
If they are to establish NSP4 in the future as a possible target protein for anti-inflammatory drugs, the scientists must now examine its function in living organisms and discover whether blocking the enzyme has adverse effects. The scientists are working with the company Novartis to answer these questions in laboratory mice. "NSP4 inhibitors could be used in diseases like chronic arthritis or inflammatory skin diseases", says Dieter Jenne, "but first we have to test the long-term effects of these substances."
Source : Max-Planck-Gesellschaft
http://www.biologynews.net/archives/2012/04/03/new_immune_defense_enzyme_discovered.html?utm_source=feedburner&utm_medium=feed&utm_campaign=Feed%3A+biologynews%2Fheadlines+%28Biology+News+Net%29
Minggu, 01 April 2012
Mipa Show 2012
Alhamdulillahirrobil 'alamin telah selesai dilaksanakannya Mipa Show Universitas Negeri Semarang tahun 2012 dengan Biologi sebagai tuan rumah. Lokasi Mipa Show di gedung D4 lantai 3 telah berjalan dengan lancar dan dihadiri oleh tamu undangan diantaranya Bapak Tyas Agung Pribadi, Syafi'i, Bapak Andin. Peserta Mipa Show terdiri dari organisasi keilmiahan jurusan Biologi, Matematika, fisika dan Kimia.
Mipa Show merupakan kegiatan yang dilaksanakan oleh MIPA Unnes yang mengarah pada English Club Departement. Mipa Show tahun ini bertemakan News and Entertain.Performance yang ditampilkan oleh masing-masing jurusan sangat menarik dan menghibur baik tamu undangan maupun peserta Mipa Show.
Harapan JSC untuk Mipa Show yaitu kedepannya terus upgrade dan mampu menciptakan karya-karya luar biasa dari Mahasiswa Mipa Universitas Negeri Semarang.
Mipa Show merupakan kegiatan yang dilaksanakan oleh MIPA Unnes yang mengarah pada English Club Departement. Mipa Show tahun ini bertemakan News and Entertain.Performance yang ditampilkan oleh masing-masing jurusan sangat menarik dan menghibur baik tamu undangan maupun peserta Mipa Show.
Harapan JSC untuk Mipa Show yaitu kedepannya terus upgrade dan mampu menciptakan karya-karya luar biasa dari Mahasiswa Mipa Universitas Negeri Semarang.
Tiny, implantable medical device can propel itself through bloodstream
Engineers at Stanford have developed a wirelessly powered, self-propelled medical device that can propel itself through the blood stream to deliver drugs, perform diagnostics or microsurgeries. Someday, your doctor may turn to you and say, "Take two surgeons and call me in the morning." If that day arrives, you may just have Ada Poon to thank.
Yesterday, at the International Solid-State Circuits Conference (ISSCC) before an audience of her peers, electrical engineer Poon demonstrated a tiny, wirelessly powered, self-propelled medical device capable of controlled motion through a fluid—blood more specifically. The era of swallow-the-surgeon medical care may no longer be the stuff of science fiction.
Poon is an assistant professor at the Stanford School of Engineering. She is developing a new class of medical devices that can be implanted or injected into the human body and powered wirelessly using electromagnetic radio waves. No batteries to wear out. No cables to provide power. "Such devices could revolutionize medical technology," said Poon. "Applications include everything from diagnostics to minimally invasive surgeries."
Certain of these new devices, like heart probes, chemical and pressure sensors, cochlear implants, pacemakers, and drug pumps, would be stationary within the body. Others, like Poon's most recent creations, could travel through the bloodstream to deliver drugs, perform analyses, and perhaps even zap blood clots or removing plaque from sclerotic arteries.
Challenged by power
"While we have gotten very good at shrinking electronic and mechanical components of implants, energy storage has lagged in the move to miniaturize," said co-author Teresa Meng, a professor of electrical engineering and of computer science at Stanford. "This hinders us in where we can place implants within the body, but also creates the risk of corrosion or broken wires, not to mention replacing aging batteries."
Poon's devices are different. They consist of a radio transmitter outside the body sending signals to an independent device inside the body that picks up the signal with an antenna of coiled wire. The transmitter and the antenna are magnetically coupled such that any change in current flow in the transmitter produces a voltage in the coiled wire — or, more accurately, it induces a voltage. The power is transferred wirelessly. The electricity runs electronics on the device and propels it through the bloodstream, if so desired.
Upending convention
It sounds easy, but it is not. Poon had to first upend some long-held assumptions about the delivery of wireless power inside the human body.
For fifty years, scientists have been working on wireless electromagnetic powering of implantable devices, but they ran up against mathematics. According to the models, high-frequency radio waves dissipate quickly in human tissue, fading exponentially the deeper they go.

This image shows the scale of the wirelessly powered, self-propelled medical device developed by electrical engineer Ada Poon at Stanford.
Low-frequency signals, on the other hand, penetrate well, but require antennae a few centimeters in diameter to generate enough power for the device, far too large to fit through all but the biggest arteries. In essence, because the math said it could not be done, the engineers never tried.
Then a curious thing happened. Poon started to look more closely at the models. She realized that scientists were approaching the problem incorrectly. In their models, they assumed that human muscle, fat and bone were generally good conductors of electricity, and therefore governed by a specific subset of the mathematical principles known as Maxwell's equations — the "quasi-static approximation" to be exact.
Poon took a different tack, choosing instead to model tissue as a dielectric — a type of insulator. As it turns out human tissue is a poor conductor of electricity. But, radio waves can still move through them. In a dielectric, the signal is conveyed as waves of shifting polarization of atoms within cells. Even better, Poon also discovered that human tissue is a "low-loss" dielectric — that is to say little of the signal gets lost along the way.
She recalculated and made a surprising find: Using new equations she learned high-frequency radio waves \ travel much farther in human tissue than originally thought.
Revelation
"When we extended things to higher frequencies using a simple model of tissue we realized that the optimal frequency for wireless powering is actually around one gigahertz," said Poon, "about 100 times higher than previously thought."
More significantly, however, her revelation meant that antennae inside the body could be 100 times smaller and yet deliver the same power.
Poon was not so much in search of a new technology; she was in search of a new math. The antenna on the device Poon demonstrated at the conference yesterday is just two millimeters square; small enough to travel through the bloodstream.
She has developed two types of self-propelled devices. One drives electrical current directly through the fluid to create a directional force that pushes the device forward. This type of device is capable of moving at just over half-a-centimeter per second. The second type switches current back-and-forth in a wire loop to produce swishing motion similar to the motion a kayaker makes to paddle upstream.
"There is considerable room for improvement and much work remains before these devices are ready for medical applications," said Poon. "But for the first time in decades the possibility seems closer than ever."
Source : Stanford School of Engineering
http://www.biologynews.net/archives/2012/02/22/tiny_implantable_medical_device_can_propel_itself_through_bloodstream.html
Langganan:
Postingan (Atom)

